Genetic testing
What are genetic diseases?
A genetic disease is a general term for a disease that develops due to an abnormality (mutation) present in an individual's genes. This type of disease is influenced by the patient's genotype and other genetic factors.
Genetic disorders are classified into many different types, but the main categories include:
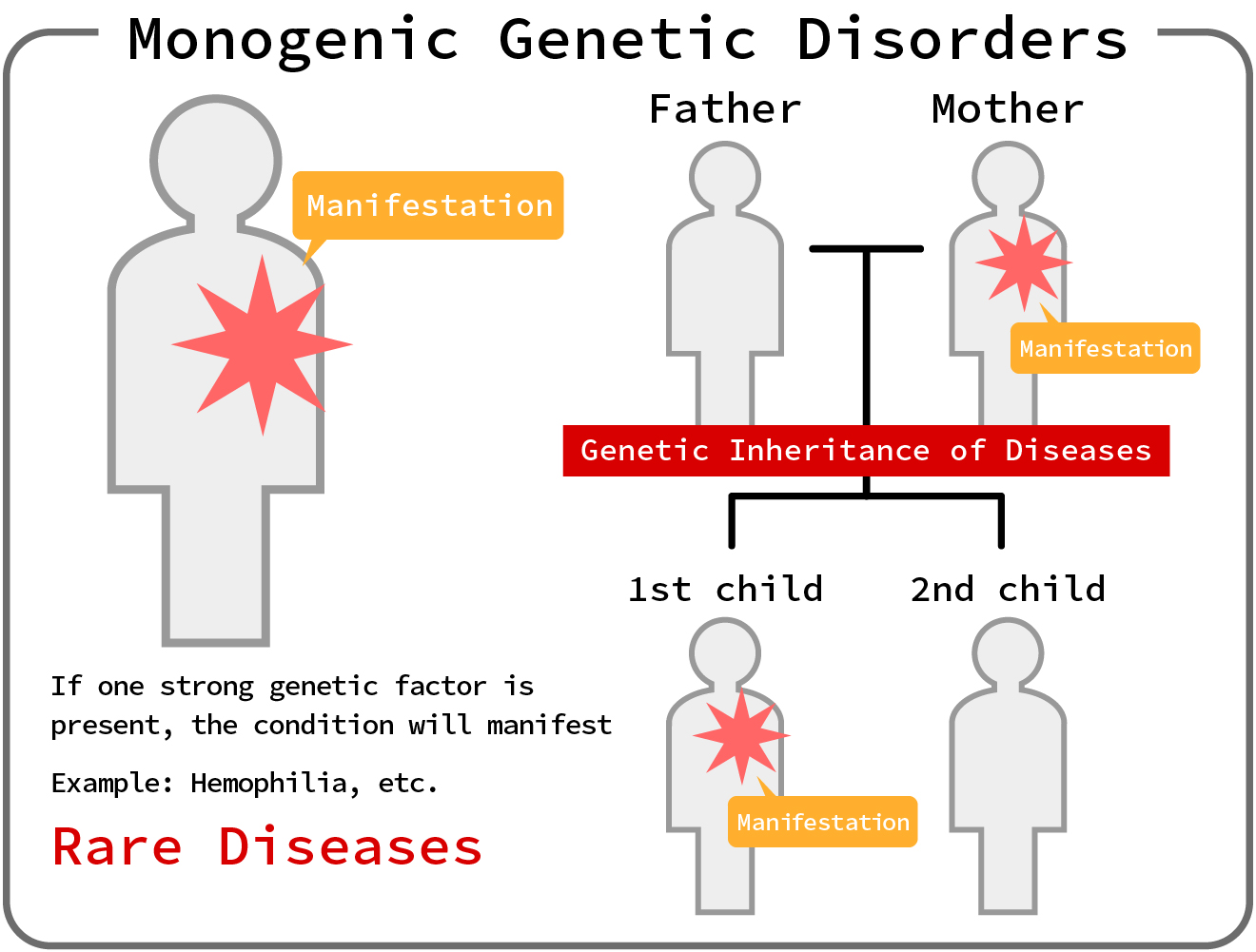
●Monogenic Disorders
Monogenic disorders are disorders caused by mutations in a single gene.
These disorders generally follow a recessive or dominant pattern of inheritance at the gene locus.
Examples include phenylketonuria and neurofibromatosis type 1.
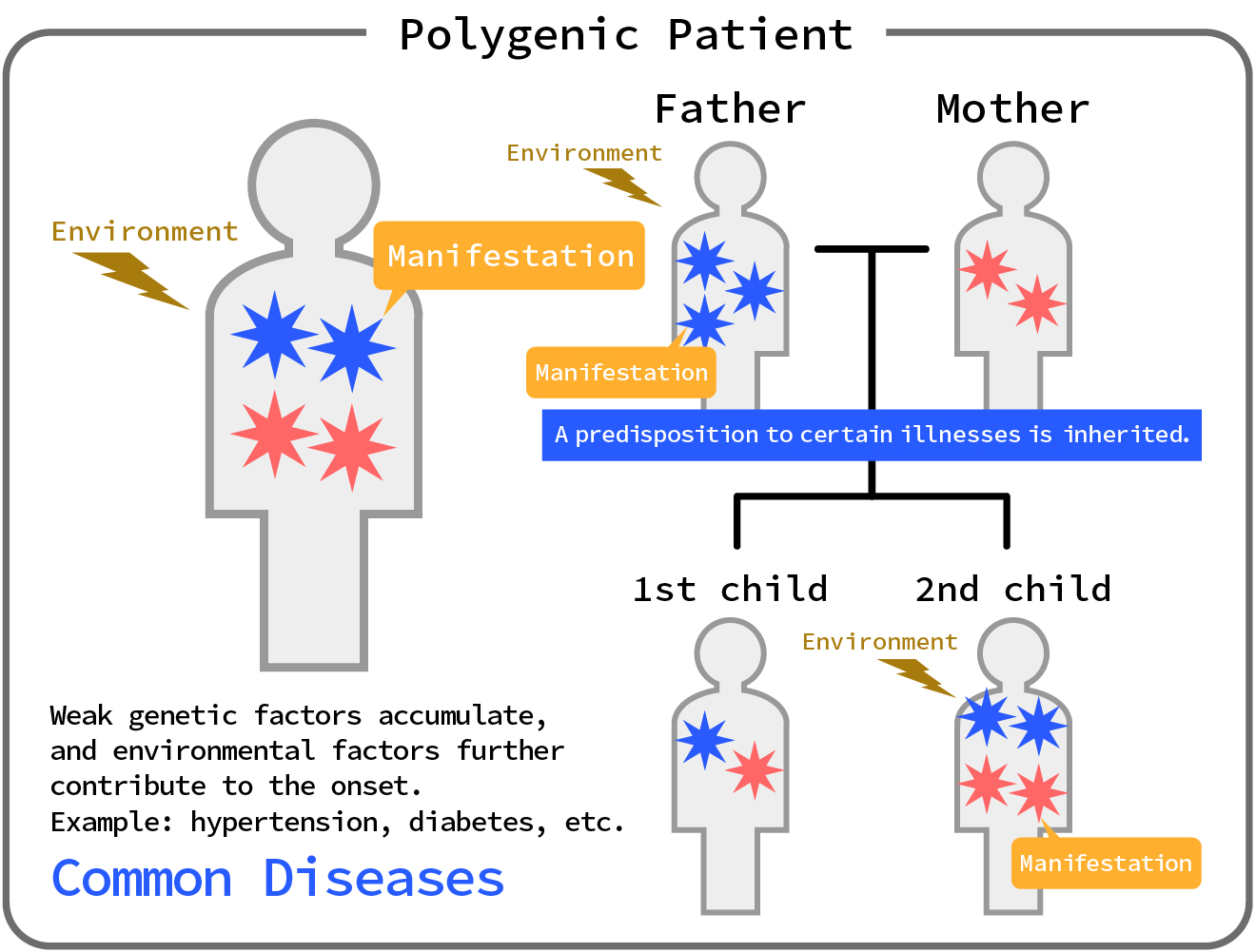
●Polygenic Disorders
Multifactorial genetic diseases are diseases caused by the interaction of multiple genetic mutations and environmental factors.
As a result, the risk of developing a disease varies depending on an individual's genetic predisposition.
Examples include type 2 diabetes and hypertension.
●Repeat Expansion Diseases
Repeat expansion disorders are caused by an abnormal expansion of a repeated sequence of DNA in a specific region of a gene, which prevents the gene from functioning normally and causes disease. These disorders include Huntington's disease and myotonic dystrophy.
●Permeability
Genetic penetrance indicates the degree to which a genetic trait affects the phenotype of an individual.
In the case of complete penetrance, all individuals carrying a gene mutation will show the corresponding phenotype, whereas in the case of incomplete penetrance, the degree of influence varies from individual to individual, and the mutation may only be present in a portion of individuals carrying the gene.
●VUS (Variant of Uncertain Significance)
VUS is a term used to indicate that the functional or clinical significance of a particular gene mutation in genetic analysis is unclear at the time of testing.
Such mutations may be difficult to provide accurate information on disease risk and diagnosis.
In addition, the assessment may change in the future, requiring long-term follow-up.
Monogenic Disorders
Single gene disorders are a group of disorders that develop due to mutations in individual genes.
These disorders are caused by genetic mutations (mutations such as deletions, substitutions, and insertions of bases) at gene loci and usually follow recessive or dominant inheritance patterns.
In the case of recessive disorders, if both alleles have a mutation, the disease caused by the mutation will develop. Conversely, if one allele is normal, the person will not develop the disease even if they have a mutation.
In the case of dominant disorders, the disease will be caused by a mutation in only one gene, so a child may inherit the disease if only one parent has a mutation.
Single gene disorders are caused by functional loss or mutation of a gene, so the loss or change of a specific function affects a specific part or function of the body.
Single-gene disorders are classified into three types based on their inheritance pattern.
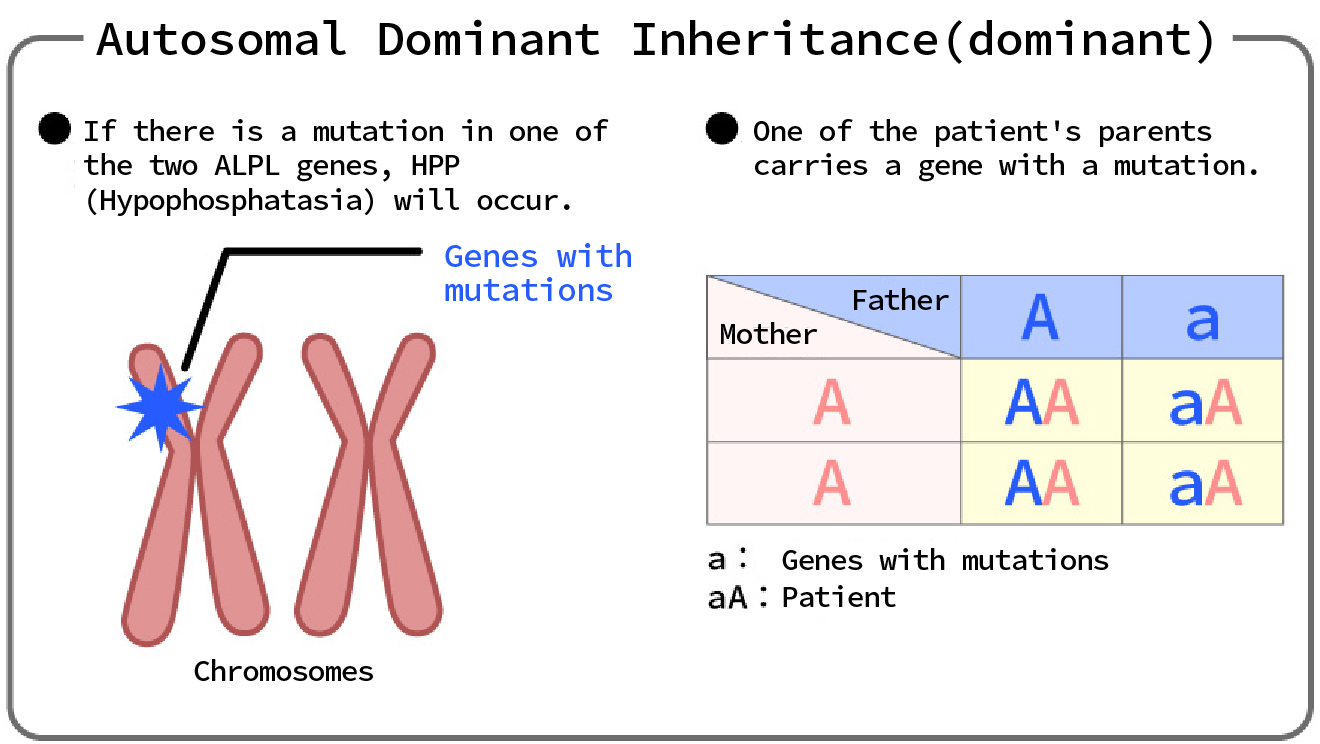
(1) Autosomal dominant (dominant) genetic disease
...If there is an abnormality in one of the paired genes on an autosomal chromosome, the disease will develop.
In dominant inheritance, only one copy of an abnormal gene (allele) is needed to cause a disease.
This means that the dominant allele has a phenotypic advantage over the recessive allele.
〔 Phenotypic Expression 〕
In dominant inheritance, the abnormal phenotype appears in both heterozygotes (Aa) and homozygotes (AA), where "A" represents the abnormal allele and "a" represents the normal allele.
〔 Patterns of inheritance 〕
Dominant disorders have a 50% chance of being transmitted from parent to child because a parent with an abnormal allele (Aa) has a 50% chance of passing it on to their child.
〔 example 〕
- Huntington's disease:
- Huntington's disease is a neurodegenerative disorder caused by dominant gene mutations that contain an abnormal expansion of CAG repeats
- Familial hypercholesterolemia:
- Mutations in the LDL receptor gene lead to abnormal cholesterol metabolism, resulting in hypercholesterolemia.
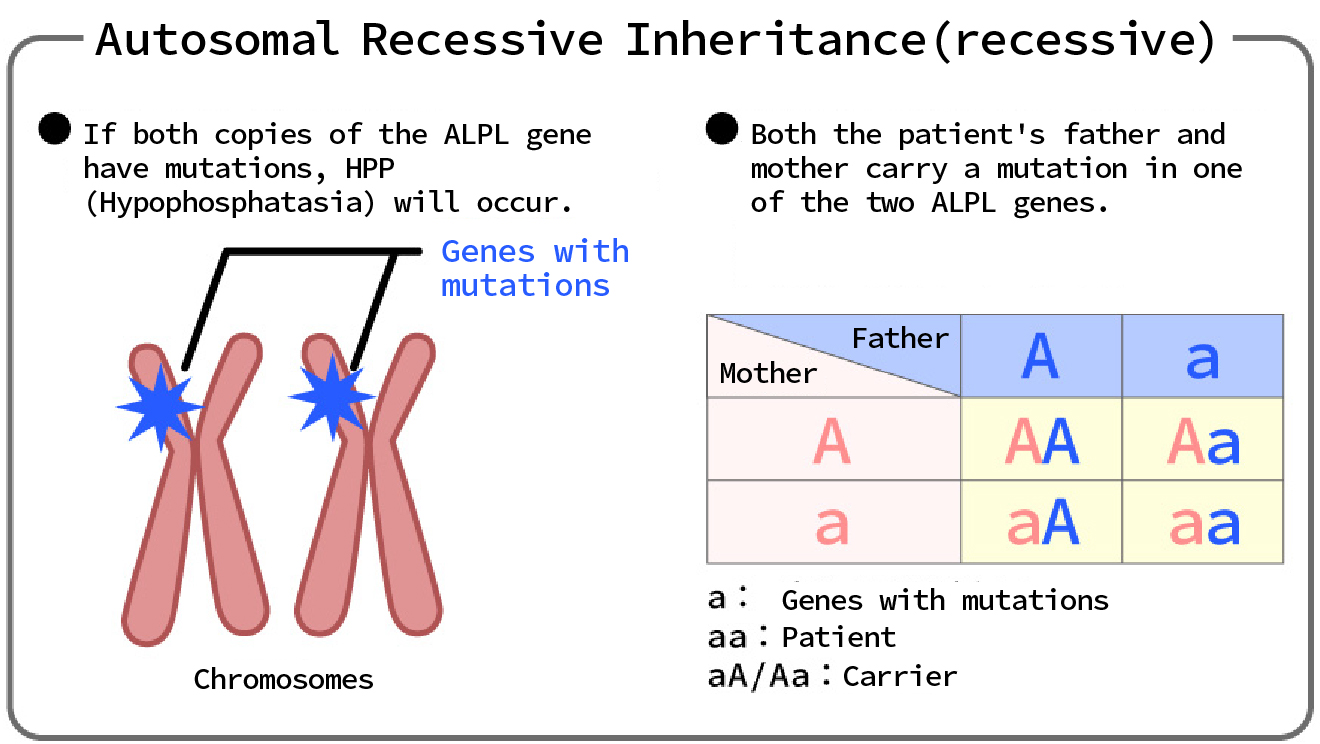
(2) Autosomal recessive genetic diseases
...If both of the paired genes on an autosomal chromosome are abnormal, the patient will develop the disease, but if only one gene is abnormal, the patient will be a carrier.
With recessive inheritance, a disease can only develop if an abnormality (mutation) is present in both copies (alleles) of a gene.
This means that the phenotype can be masked by the normal allele.
〔 Phenotypic Expression 〕
In recessive inheritance, an abnormal phenotype only appears when the individual is homozygous (aa), where "a" represents the abnormal (mutated) allele and "A" represents the normal allele.
Heterozygotes (Aa) show a normal phenotype, but these individuals are called carriers and may pass on the abnormal gene to future generations.
〔 Patterns of inheritance 〕
For recessive genetic diseases, if both parents are carriers, the child has a 25% chance of developing the disease.
This is because in the combination of carriers (Aa x Aa), 25% of children will be homozygous (aa).
Also, 50% will be carriers (Aa) and 25% will be normal (AA).
〔 example 〕
- Cystic fibrosis:
- Mutations in the CFTR gene cause abnormal mucus secretion, leading to serious problems in the lungs and digestive system.
- Phenylketonuria (PKU):
- Mutations in the PAH gene impair phenylalanine metabolism, causing intellectual disability and developmental delay.
Source:https://hpp-life.jp/about/genetics
(3) X-linked genetic diseases
...There is an abnormality in the X chromosome, which is a sex chromosome.
- ●Inheritance related to sex chromosomes:
- Single-gene disorders may also be linked to the sex chromosomes (X and Y).
These disorders have different inheritance patterns depending on the gender. - ●X-linked recessive inheritance:
- In X-linked recessive inheritance, the abnormal (mutated) gene is located on the X chromosome.
This type of disorder is more common in males.
〔 Phenotypic Expression 〕
Males (XY) have only one X chromosome, so they have no normal alleles and only one abnormal allele (Xa) is needed to cause the disease.
Females (XX) have two X chromosomes, so they only need two abnormal alleles (XaXa) to cause the disease.
Those with only one abnormal allele (XaX) are carriers, but usually do not show symptoms.
〔 Patterns of inheritance 〕
When a carrier female (XaX) and a normal male (XY) are paired, there is a 50% chance that a boy will have the condition and a 50% chance that a girl will be a carrier.
When an affected male (XaY) and a normal female (XX) are paired, all girls will be carriers and all boys will be normal.
〔 example 〕
- hemophilia:
- A disease in which bleeding becomes difficult to stop due to a deficiency of blood clotting factors.
- Duchenne muscular dystrophy:
- A disease that causes progressive muscle weakness.
Overt (dominant) and recessive (recessive) inheritance are important concepts in the development of single-gene disorders.
Overt (dominant) inheritance requires only one abnormal gene to cause the disease, whereas recessive (recessive) inheritance requires both genes to be abnormal.
Understanding these differences provides a foundation for genetic disease risk assessment, genetic counseling, and treatment development.
Genetics or mutation
In the case of monogenic diseases, whether the onset is due to a new (de novo) mutation or hereditary causes varies by disease. In general, it is believed that hereditary causes are more prevalent.
In cases where the condition is inherited, the patient is more likely to inherit it from a parent who has the mutation. In cases where the condition is recessive, children can develop the disease even if only one parent has the mutation. In cases where the condition is dominant, children are more likely to develop the disease if only one parent has the mutation.
However, de novo mutations can also cause disease. De novo mutations are not usually inherited from parents, but rather occur in an individual's germ cells or during fetal development. Although de novo mutations can cause disease, these diseases are usually genetically unlikely to be recurring, making risk assessment based on family history difficult.
Thus, many monogenic disorders are primarily due to inheritance, and de novo mutations are relatively rare, but may vary depending on the specific disorder and mutation.
Disease Examples
- ●Achondroplasia
- Achondroplasia is a genetic disorder characterized by abnormalities in cartilage that affect skeletal morphology and growth.
It is caused by mutations in the FGFR3 gene, which causes abnormal gene function and excessive suppression of bone growth, resulting in short stature and shortened limbs.
- ●Duchenne Muscular Dystrophy (DMD)
- Muscular dystrophies refer to a group of disorders characterized by muscle degeneration and weakness.
Duchenne muscular dystrophy is caused by a mutation in the DMD gene on the X chromosome. This mutation results in insufficient production of dystrophin, a protein that supports muscles, resulting in impaired muscle structure and function.
- ●Huntington's Disease (HD):
- Huntington's disease is a degenerative disorder of the central nervous system that causes impaired motor and cognitive functions due to the progressive death of nerve cells.
This disease is caused by mutations in the HTT gene and shows a dominant inheritance pattern.
- ●Congenital Myasthenic Syndromes (CMS):
- Congenital polymyositis is a group of disorders caused by dysfunction of the neuromuscular junction.
These disorders are caused by mutations in genes involved in the release and reception of neurotransmitters.
- ●Phenylketonuria (PKU):
- Phenylketonuria is a metabolic disorder caused by abnormalities in phenylalanine metabolism.
The disease is caused by a deficiency in the phenylalanine hydroxylase (PAH) enzyme.
- ●Cystic Fibrosis (CF)
- Cystic fibrosis is a genetic disease that primarily affects organs such as the lungs, pancreas, and digestive tract. The disease is caused by mutations in the CFTR gene. The CFTR gene codes for a protein that regulates chloride ion (anion) transport, and mutations in the gene disrupt normal chloride ion transport.
Symptoms of CF include chronic respiratory infections, reduced lung function, digestive tract problems (such as pancreatic insufficiency and intestinal blockages), difficulty gaining weight, and insufficient salt excretion. The disease shows a recessive inheritance pattern, and if both parents carry the mutation, their children may develop CF. The diagnosis of CF is based on genetic testing and clinical symptoms, and early detection and treatment are important.
CF now has advanced treatments, including symptomatic and drug therapy, which can improve the quality of life of patients. However, there is still no cure for the disease, and patients require ongoing management and treatment throughout their lives.
Diagnosis and Treatment
Diagnosis of single-gene disorders is made through family history and genetic testing. The cooperation of a genetic counselor or clinical geneticist is also important. Treatment varies depending on the disease and symptoms, but generally aims to alleviate symptoms and slow progression. For example, physical therapy and certain drug therapy may be used. In addition, PGT-M allows embryos created by IVF to be examined for mutation status and transferred (pregnancy).
Single-gene disorders are discovered in the following ways:
 Family history investigation
Family history investigation- First, we investigate how the disease spreads within families. It is important to find out how many generations a particular disease has been transmitted within a family. Based on family history, characteristic inheritance patterns may be found.
 Genetic analysis and gene mapping
Genetic analysis and gene mapping- To identify disease-causing genes, genetic analysis or gene mapping is used, which involves DNA sequencing and analysis with genetic markers. The goal is to identify which mutations in specific genes are associated with a particular disease.
 Genetic counseling
Genetic counseling- Genetic counseling is the process of providing information related to genetic risk and genetic test results, including family history and genetic risk assessment, test interpretation, and disease risk and management information.
 Functional analysis
Functional analysis- When a mutation in a particular gene is found, functional analysis is performed to understand how the function of the gene is affected, which can include in vitro and in vivo experiments and analyses in cell and animal models.
 Clinical Trials
Clinical Trials- When mutations in a particular gene are identified as being associated with a particular disease, clinical trials may be conducted to find ways to treat or prevent that disease.
Through these steps, it is confirmed that a specific gene mutation is associated with a specific disease, and research is then conducted to understand the disease and develop a treatment for it.
Latest research trends
Research into single-gene diseases is evolving with innovative approaches, such as the development of gene editing technology and new ssibilities for gene therapy.
This will hopefully lead to the development of new therapies that treat the genetic causes. In addition, rather than directly treating the cause of the disease, it is possible to examine embryos created through IVF (PGT-M) and select embryos that are not affected by the disease for implantation and pregnancy.
summary
Monogenic diseases are diseases caused by mutations in individual genes and are diagnosed through genetic analysis and family history.
Treatments vary depending on the disease and symptoms, but generally aim to alleviate symptoms and slow progression. The latest research trends are expected to lead to the development of new treatments and strategies to improve the quality of life for patients.
[Quoted site]]
https://www.invitra.com/en/monogenic-disorders/
https://www.msdmanuals.com/ja-jp/home/01-知っておきたい基礎知識/遺伝学/単一遺伝子疾患の遺伝?ruleredirectid=464
https://www.msdmanuals.com/ja-jp/professional/24-その他のトピック/遺伝医学の一般原則/単一遺伝子の異常?ruleredirectid=465
Polygenic diseases: genetic complexity and lifestyle abyss
Polygenic diseases are a very interesting and complex area of genetics where multiple genes are involved.
Unlike monogenic diseases where a single gene mutation causes the disease, polygenic diseases are caused by many gene mutations interacting with each other to contribute to the onset of the disease.
This page focuses on lifestyle-related diseases such as hypertension and diabetes, and provides a detailed explanation of advanced genetic research techniques such as genome-wide association studies (GWAS), single nucleotide polymorphisms (SNPs), linkage analysis, and Mendelian randomization.
Understanding Polygenic Disorders
Common examples of polygenic diseases include cardiovascular disease, diabetes, obesity, depression, and schizophrenia.
These diseases are considered multifactorial because they are influenced by both genetic and environmental factors, and unlike monogenic diseases, they are caused by the cumulative effect of many genes.
Lifestyle-related diseases: High blood pressure and diabetes
〔 High Blood Pressure 〕
Hypertension is a chronic condition characterized by persistently high blood pressure in the arteries. It is a major risk factor for cardiovascular diseases such as heart attack and stroke.
- Genetic factors:
- Many genetic loci are associated with hypertension, including genes that regulate physiological processes such as renal sodium handling, vascular tone, and the renin-angiotensin system.
- Environmental factors
- Lifestyle habits such as a high-salt diet, lack of exercise, and stress significantly affect the risk of high blood pressure.
〔 Diabetes mellitus 〕
Diabetes, particularly type 2 diabetes, is a metabolic disease characterized by hyperglycemic states due to insulin resistance or insufficient insulin secretion.
- Genetic factors
- Several genetic variants that affect insulin secretion and glucose metabolism have been identified, often in genes involved in pancreatic β-cell function and the insulin signaling pathway.
- Environmental factors
- Diet, obesity, and lack of exercise are important environmental factors in the development of type 2 diabetes.
Single nucleotide polymorphisms (SNPs)
〔 What is a SNP? 〕
Single Nucleotide Polymorphism (SNP) is the most common form of genetic variation in genes and refers to a difference in a single base (A, T, C, G) that makes up DNA at a specific position in the genome. The human genome contains about 3 billion base pairs of DNA, among which about 10 million SNPs are known. These variations are an important factor in forming genetic diversity between individuals.
●How to detect SNPs
Technologies for detecting SNPs are evolving rapidly. Common methods include:
- 1) DNA sequencing
- Next-generation sequencing (NGS) technology is used for fast and accurate detection of SNPs across large genomes.
- 2) Genotyping array
- Specific SNP-targeted array technologies are used to detect many SNPs at once, and these arrays are cost-effective and suitable for large population studies.
●SNP database
The detected SNPs are registered in databases. Representative databases include the following:
- 1) dbSNP
- This is a database of SNPs provided by NCBI, and contains information on millions of SNPs.
- 2) 1000 Genomes Project
- This project comprehensively covers human genetic diversity and makes SNP data collected from different populations around the world publicly available.
〔 Classification of SNPs 〕
SNPs are classified based on the gene function at their location as follows:
1) Coding region SNPs
- ●Synonymous SNPs
- A mutation that does not affect the amino acid sequence coded by a gene. The encoded amino acids are coded for by three bases (codons), which code for 20 different amino acids. Therefore, there are multiple combinations of bases that code for one amino acid. For example, the codons GAA and GAG both code for glutamic acid. Therefore, even if the third base mutates from A to G, the same amino acid is coded for. Therefore, there is no difference in the coded amino acid sequence.
- ●Nonsynonymous SNPs
- A mutation that changes the amino acid sequence encoded by a gene. Nonsynonymous SNPs are further divided into missense mutations (where one amino acid coded for by a gene is replaced by another) and nonsense mutations (where an amino acid is replaced by a stop codon, terminating the encoded amino acid sequence).
2) Non-coding region SNPs
Mutations located in regions that regulate the location and timing of gene expression (promoters and enhancers) or intron regions. They can affect gene splicing and the efficiency of transcription.
〔 Functional effects of SNPs 〕
SNPs have various effects on the functions of genes and proteins, including the following:
- 1) Regulation of gene expression
- SNPs located in regulatory regions can alter gene expression levels through transcription factor binding and changes in chromatin structure.
- 2) Changes in protein function
- Nonsynonymous SNPs alter the amino acid sequence of a protein, thereby affecting its function and stability.
- 3) Changes in splicing
- SNPs located at intron-exon boundaries can alter splicing patterns.
〔 SNP research and applications 〕
1) Disease-related research
- ●GWAS
- Research to identify genetic variants associated with disease using SNPs. Finding the association between a specific SNP and a disease clarifies the onset mechanism and risk factors of the disease. GWAS will be explained in more detail in the next section.
- ●Disease risk prediction
- When the presence of a particular SNP is known to increase the risk of a particular disease, for example through GWAS results, it can be used to assess an individual's risk.
- ●Functional studies
- Investigating how differences in specific SNPs affect gene function can be used to deepen our understanding of disease pathology and elucidate the mechanisms underlying disease development.
2) Personalized medicine
Using SNP information, it is possible to select the most appropriate treatment and drug for each individual patient. For example, SNPs in genes involved in drug metabolism can affect the efficacy and side effects of drugs, helping to determine the appropriate dosage for each patient.
It also impacts risk assessment and progression prediction for polygenic diseases.
3) Evolution and population genetics
SNP data are used to study genetic variation among different populations,which can shed light on human evolution, migration patterns, and genetic relationships between populations.
Genome-wide association study (GWAS)
GWAS are studies that scan the entire genome for SNPs associated with a particular disease or trait. By comparing the genomes of people with and without the disease, genetic variants that contribute to disease risk can be identified.
GWAS collects DNA samples from large cohorts and analyzes hundreds of thousands to millions of SNPs using high-throughput genotyping.
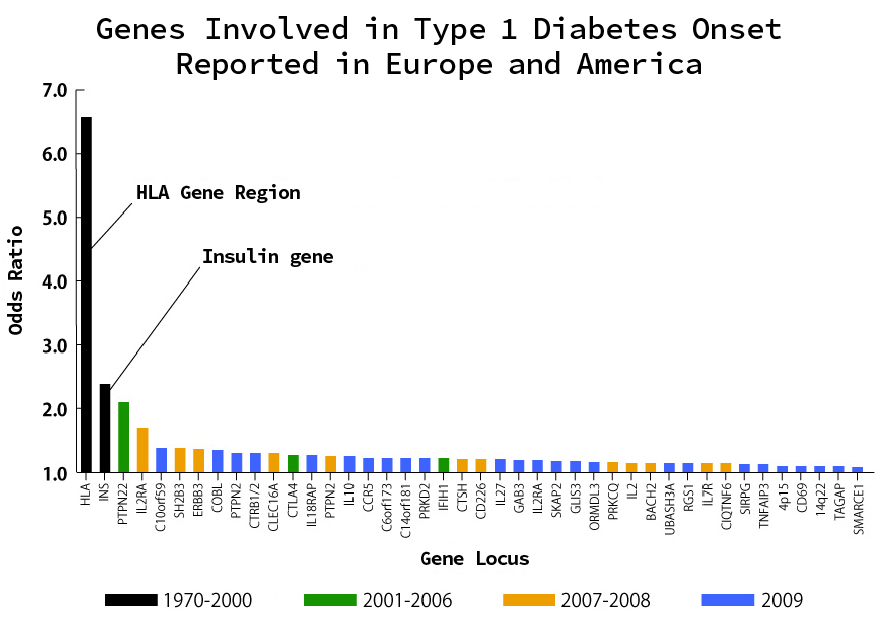
As a result, many risk loci related to hypertension and diabetes have been identified by GWAS. Although the contribution of each locus identified by GWAS to the disease is small, their accumulation contributes to the development of polygenic diseases.
Linkage analysis
Linkage analysis is a method to create a genetic map and identify disease-causing genes by examining the inheritance patterns of marker genes (SNPs) within families.
- ●Principle
During meiosis, which produces sperm and eggs, genetic recombination occurs between homologous chromosomes.
The frequency of recombination of genes that are physically close to each other is lower than that of genes that are far apart, making them more likely to be inherited together.
Based on this frequency, the distance between genes is estimated and a genetic map is created.
From there, marker genes that are closer (more likely to be inherited together) are found to narrow down the causative gene, and ultimately the causative gene is identified.
Application to polygenic diseases: Although linkage analysis is primarily used for monogenic diseases, it can also be useful for identifying genomic regions that contribute to polygenic traits.
However, due to the increased complexity of the analysis, it is not as effective as GWAS for diseases with complex inheritance patterns, such as polygenic diseases.
Mendelian randomization
Mendelian randomization is a method that uses genetic variants (SNPs) to infer causal relationships between risk factors and disease in observational studies.
- ●Concept
- When investigating the effectiveness of a drug, patients can be randomly assigned to either a group that receives the drug or a group that does not, allowing the causal relationship between the drug and the treatment outcome to be determined.
However, polygenic diseases are influenced by both environmental and genetic factors, making it difficult to determine the causal relationship between a specific risk factor and the disease.
Therefore, we use the fact that genes are randomly distributed from parents to children to estimate the causal relationship between risk factors and the disease.
This mimics the randomization process of clinical trials.
- ●Application
- It helps to clarify whether the associations seen in observational studies are causal.
For example, let's say you want to estimate the causal relationship of high body mass index (BMI) causing diabetes. First, find a gene (SNP) that is not related to diabetes but is correlated with BMI. You can estimate the causal relationship by evaluating the proportion of people with this gene and the proportion of people with diabetes.
An integrated approach
Modern research combines SNP analysis, GWAS, linkage analysis, and Mendelian randomization to provide a comprehensive understanding of polygenic diseases.
These integrated approaches help elucidate the genetic architecture of diseases and identify potential therapeutic targets,thereby aiming to target causal rather than symptomatic treatments.
Summary
Lifestyle-related diseases such as hypertension and diabetes are polygenic diseases that represent a complex interplay between genes and the environment.
Advanced genetic research techniques such as SNP analysis, GWAS, linkage analysis, and Mendelian randomization are powerful tools to unravel this complexity.
Understanding these diseases at the molecular level opens the door to personalized medicine based on an individual's genetic makeup.
Exploring the depths of polygenic disease helps us understand the incredible complexity of human health and disease, and highlights the need for continued research and innovation in genetic and environmental factors.
Inquiries │
We accept inquiries by email. Tokyo · Osaka │ O.G.M.S.

Prenatal Test for Determining Fetal Sex
english_help@ogms.biz
The email address may vary depending on the content of your inquiry. Please check the mail address on each service page.
There may be a delayed reply to inquiries received at night, weekends/holidays, winter and summer break.
It may take some time to reply depending on the nature of the inquiry.